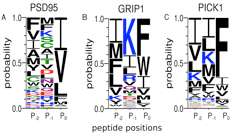
Protein-protein/peptide interactions are involved in wide range of cellular processes. Among these, interactions with disordered peptide segments, often found within longer regions of disorder in proteins, are highly interesting. They typically undergo a binding-induced folding transition upon contact with a target molecule such that a specific structure is assumed. It is not uncommon, however, that significant conformational diversity persists even after binding. Disordered regions in proteins play pivotal roles in controlling cellular signaling networks, protein subcellular localization, protein degradation, and post-translational modification. Remarkably, a recent estimate suggests that as much as around 40% of all links in protein interaction networks are due to binding of short peptide segments of around 3–10 amino acids in length to protein domains. Recently, we developed and tested a theoretical framework for exploring, in an efficient and representative way, the combined sequence and conformational space of peptides interacting with a given peptide-binding pocket. Our future interests are to further improve the current method and also investigate the molecular details of peptide aggregation that may play significant role in many neurodegenerative diseases such as Alzheimer’s and Parkinson’s diseases.