PlantG4DB: A Database of G-Quadruplex Sequences in Plants
How to use
Browse PlantG4DB is an online, open source database to predict G-quadruplexes in plants. Its highly user-friendly interface allows users to browse for GQSes in different plant species available via selecting/clicking any plant species. The page displays unique identifier for each GQS sequence (G4 identifier), organism name (Plant), GQS type (G2L1, G2L1-2, G2L1-4, G3L1-3 and G3L1-7), chromosome location and start position on that particular chromosome. User can select any of G4 identifier to display more details related to it. 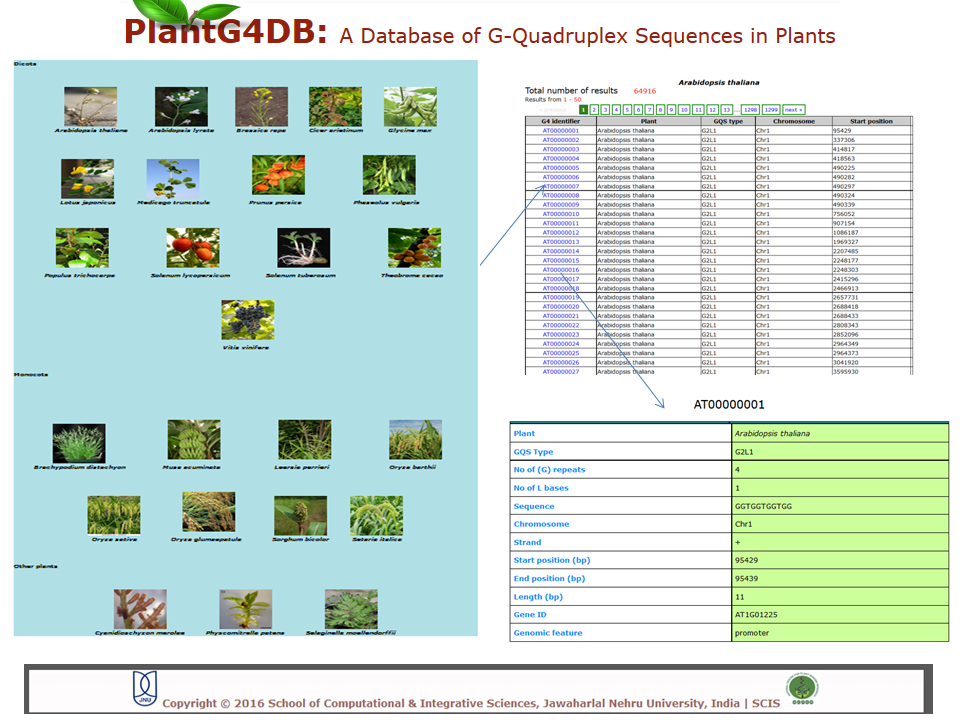 Search Under this tab users can choose from three options, including Genome search, gene search and orthologous search. Genome Search Under this tab users can search a selected plant genome for a specific type of GQS within a particular chromosome and/or location within the genome. Users can also specify strand information, position on chromosome, length of the GQS and number of G-repeats. The result displays unique identifier for each GQS sequence (G4 identifier), organism name (plant), GQS type (G2L1, G2L1-2, G2L1-4, G3L1-3 and G3L1-7), chromosome location, strand information, start position on that particular chromosome, length of GQS sequence, number of times G2 or G3 are repeated, genomic feature and gene identifier from the selected plant species. User can then select any of G4 identifier to display more details related to it. 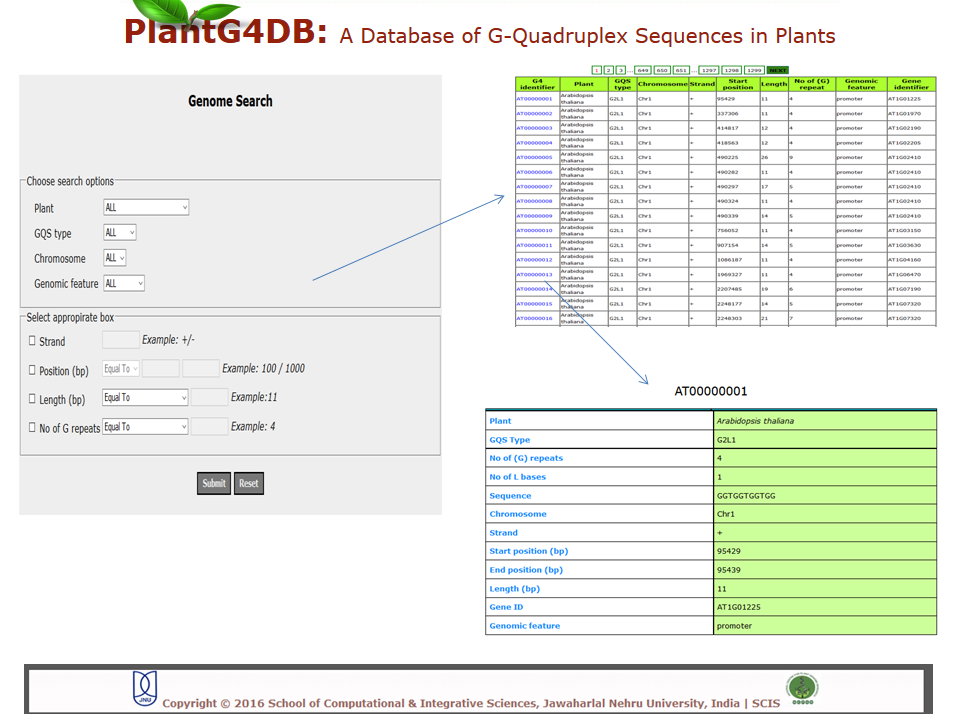 Gene Search Under this search parameter user can search for a set of genes from a particular plant by selecting the organism name and providing the list of gene identifiers. This will display a list of gene identifiers containing GQSs. The information is displayed for each gene identifier along with the type of GQS sequence present, its position on chromosome and length of GQS sequence. This option also displays the information on the genomic location (promoter, gene, intron, exon, CDS, mRNA) for each G4 identifier. 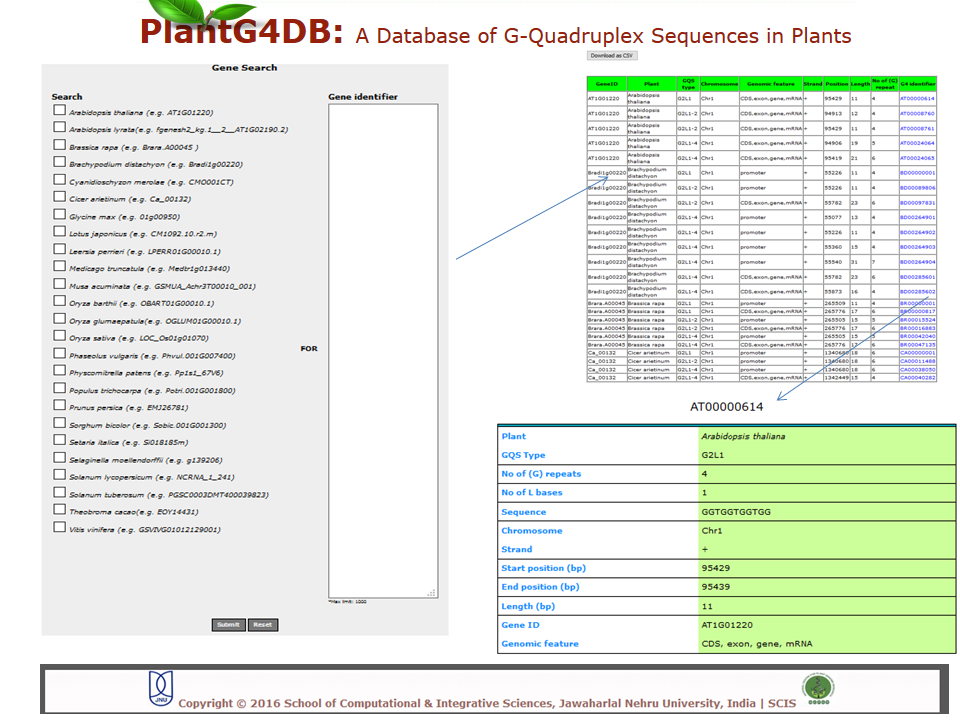 Orthologous Search This form helps users to search orthologous gene between Arabidopsis thaliana/Oryza sativa (as reference) with any other plant species. After selecting appropriate plant species and GQS type, clicking on Submit tab will display list of orthologous genes of chosen plants with details of GQSes associated with them. Further, GQSes conserved between Arabidospis/rice and any/all of the monocots/dicots can also be searched via selecting the appropriate search option. User can either select any of G4 identifier to display details of the Identifier. 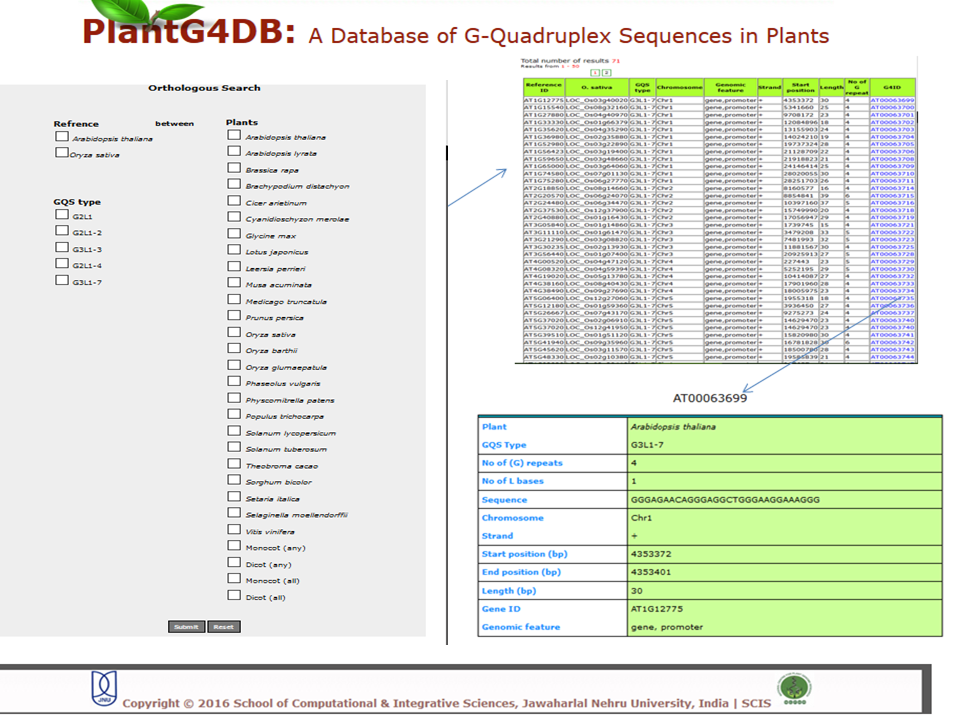 Tools This tool can help prediction of GQSes in the given input nucleotide sequence/s. Users can paste nucleotide sequences in FASTA format in the text area or upload file via browse option. Load example link loads the example data in text area (enter sequence below in Fasta format). Users can optimize the search options stringency, stem length (1-5), minimum loop length (1-50 bp), maximum loop length (1-50 bp) and search in reverse strand of the input sequence. After selecting the desired parameters, clicking on Submit button will display the GQSes present the input nucleotide sequence. 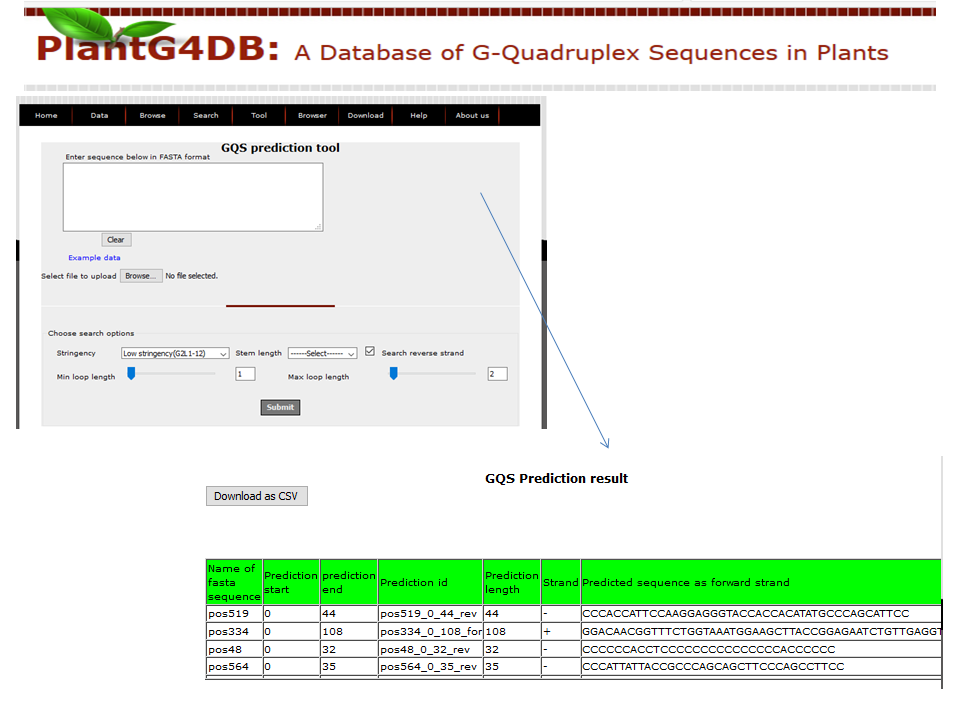 Browser A genome browser view for all the plants has also been implemented in the database to provide graphical interface for display of GQS information from the PlantG4DB. It enables researchers to visualize and browse entire genomes with identified GQSes along with annotated, including gene, CDS, exon and introns. Users can zoom in and scroll over chromosomes, showing the location of GQSes with respect to genes annotated for selected plant species. 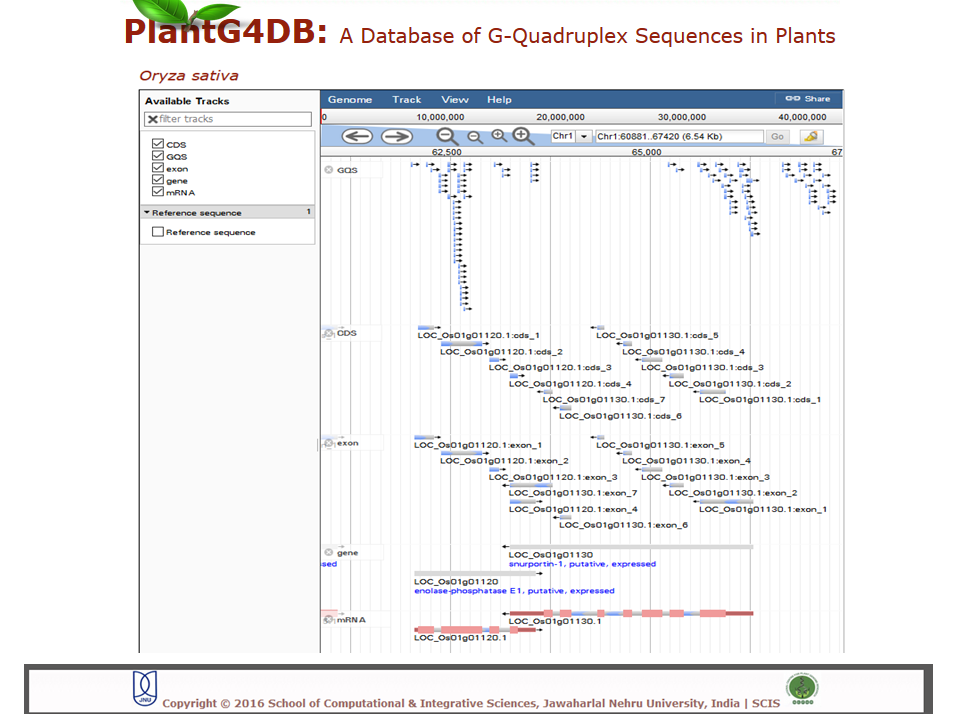 Download Users can download all the GQSes predicted for a particular plant species in .xls format by selecting the species under this tab.  |