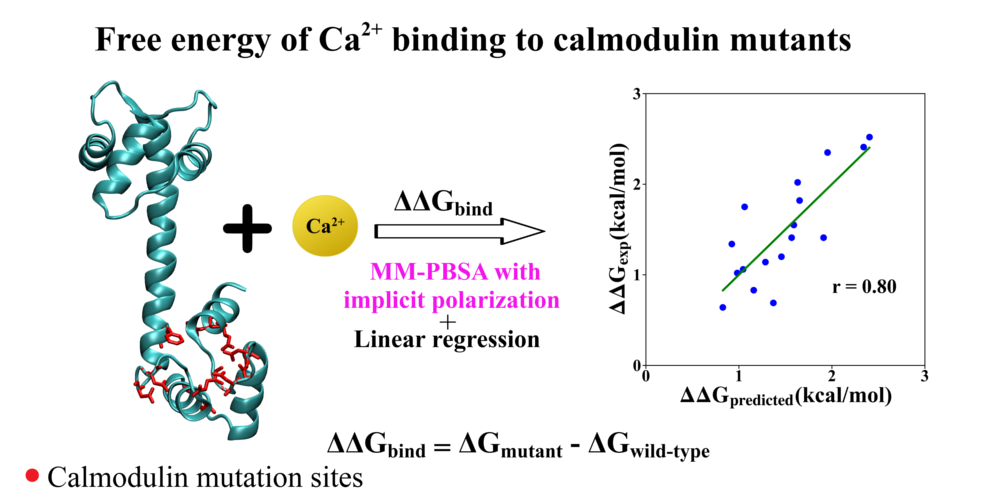
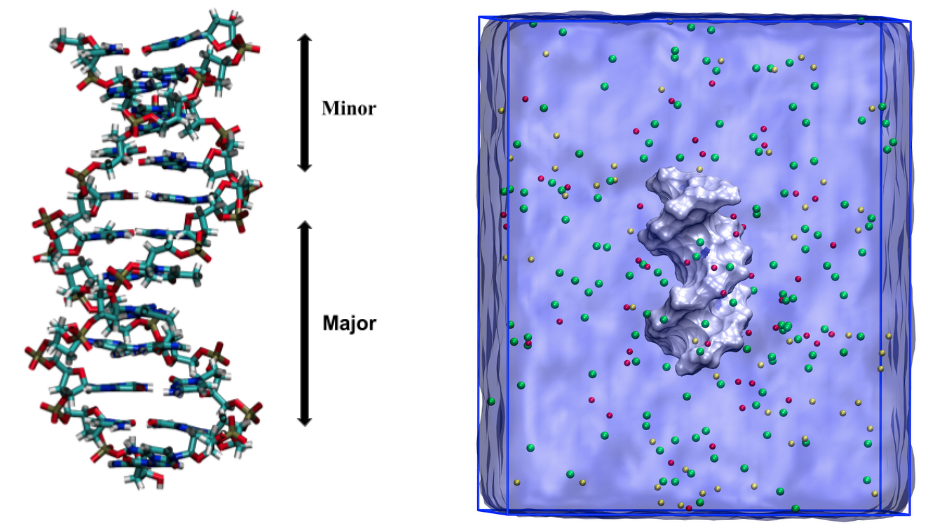
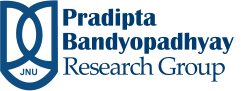Development of enhanced Monte Carlo simulation techniques to explore the energy landscape of water clusters and biomolecules
We have developed an efficient Monte-Carlo-based method, Temperature Basin Paving, for the exploration of complex energy landscapes. We have successfully applied this method to different sizes of water clusters and small RNA hairpins and continuously optimizing this method for complex systems.
Properties of RNA with non-Watson-Crick base pair
Non-Watson-Crick base pairs are the basic necessity for different three-dimensional folds of RNAs and the activity of different Ribozymes. These folds make the interaction of RNAs with proteins, other molecules, and ions possible. Non-Watson-Crick base pairs always need some external factors like metal ions, other ribosomal strand or proteins for stabilization. In the current work, we are analyzing different aspects of the stabilization mechanism of non-Watson-Crick base pairs by these external factors.
Development of a model of the cytoplasm of a bacterial cell and understanding of diffusion and hydrodynamics
In this work, we have developed a computational model of E. coli cytoplasm to study the diffusive motion of proteins inside the cell. The model contains the most abundant proteins present E. coli cytoplasm that has been coarse-grained in the form of a collection of spheres. The presence of a large number of particles provides large volume exclusion and hydrodynamic interactions which in turn affect the diffusivity of proteins inside the cell. In this work, we are trying to determine the effect of crowding and hydrodynamic interactions on the diffusive motion of proteins.
Development of Random Walk model to study anomalous diffusion
Subdiffusion is ubiquitous in a crowded heterogeneous environment, but the exact cause of subdiffusion is still debatable. In this work, we are trying to understand the cause of subdiffusion in a crowded and heterogeneous environment using random walk models. By developing different models, we are working to understand the cause of transient and pure anomalous diffusion with constant diffusion exponent at all time scales.
Effect of molecular crowding on biomolecular systems
Living cells have vast varieties of molecules like nucleic acid, protein, small messenger molecules, osmolytes, carbohydrates, and insoluble molecules. These molecules are known to occupy a significant fraction of cellular volume(20-40%) and are referred to as molecular crowders. Molecular crowding has large quantitative effects on the structure, thermodynamics, and function of nucleic acids and proteins by excluding volume and interacting with them. It has been shown that molecular crowding affects properties of nucleic acid like melting temperature, hybridization, structural preference, diffusion, etc. In this work, we are trying to understand the structural, hydration, and thermodynamics of the canonical form of DNA in a crowded environment. One of the ways to understand DNA stability is to understand the thermodynamics of its hydration, which we are currently focused to look at.
Effect of salt electrostatics on like-charged protein-protein binding
Here, we are trying to explore the effect of salt ions on the like charged protein-protein binding. How does salt ion concentration in the system affects the like-charged protein-protein binding and what roles are played by salt ions near the binding interface which determines the free energy change of complex formation, are the questions we are trying to address.